 |
biomcmc-lib
0.1
low level library for phylogenetic analysis
|
 |
biomcmc-lib
0.1
low level library for phylogenetic analysis
|
Reads tree files in nexus format and creates a vector of topologies. More...
#include "newick_space.h"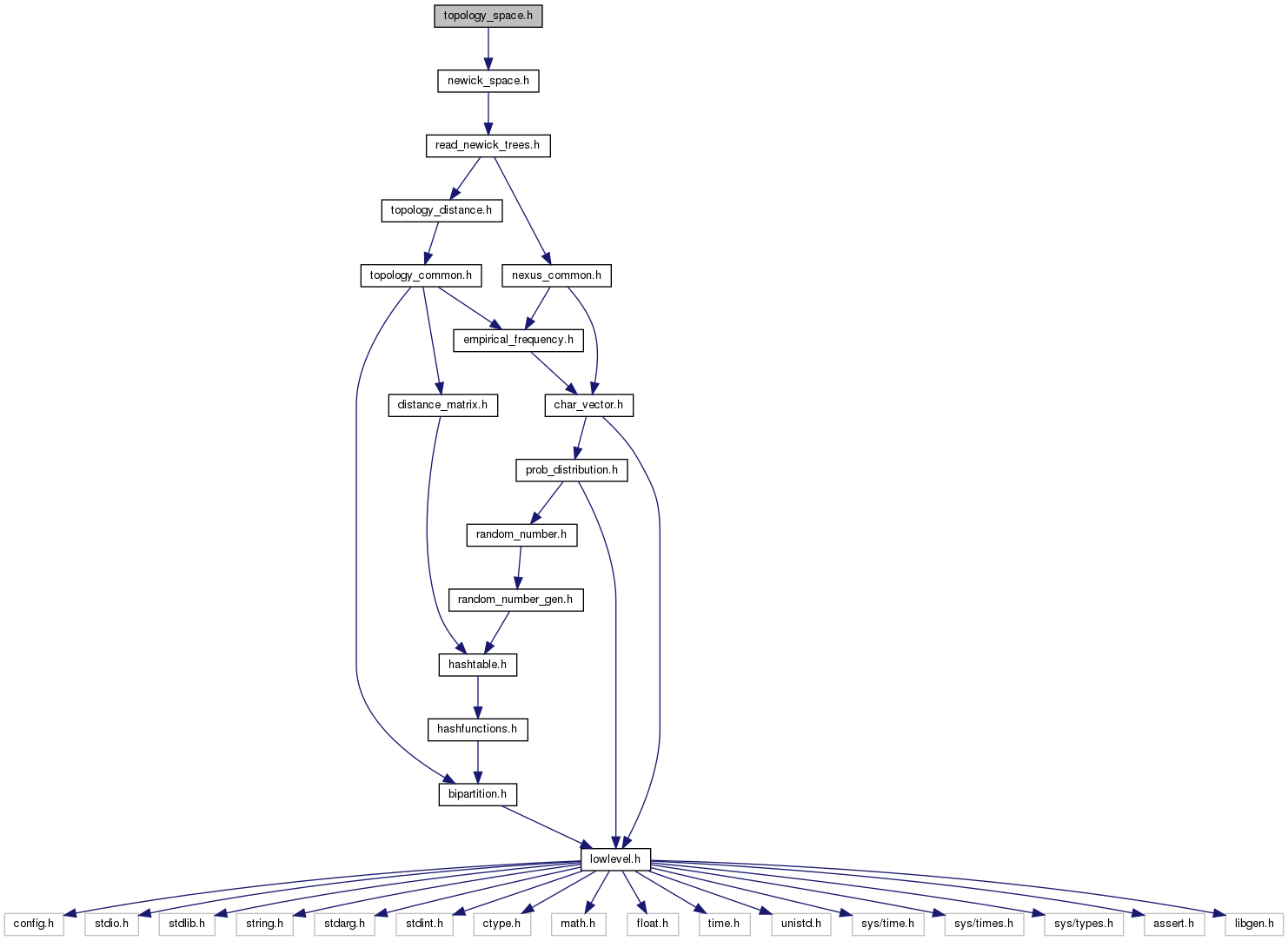
Go to the source code of this file.
Data Structures | |
| struct | topology_space_struct |
| Collection of topologies from tree file. When topologies have no branch lengths we store only unique topologies. More... | |
Typedefs | |
| typedef struct topology_space_struct * | topology_space |
Functions | |
| void | add_string_with_size_to_topology_space (topology_space *tsp_address, char *long_string, size_t string_size, bool use_root_location) |
| Read tree in newick format until char string_size, returning updated topolgy_space. Auxiliary for python module. More... | |
| void | add_topology_to_topology_space_if_distinct (topology topol, topology_space tsp, double tree_weight, bool use_root_location) |
| Add topology to topology_space only if unrooted version is distinct, updating freqs, trees[] etc. Aux for python module. | |
| topology_space | read_topology_space_from_file (char *seqfilename, hashtable external_taxhash, bool use_root_location) |
| Read tree file and store info in topology_space_struct with possible external hashtable to impose the leaf ordering. | |
| topology_space | read_topology_space_from_file_with_burnin_thin (char *seqfilename, hashtable external_taxhash, int burnin, int thin, bool use_root_location) |
| lower level function where we can specify burnin and thinning factor, in iterations | |
| void | merge_topology_spaces (topology_space ts1, topology_space ts2, double weight_ts1, bool use_root_location) |
| merge trees from two topology_space objects, assuming names hashtable is the same | |
| void | sort_topology_space_by_frequency (topology_space tsp, double *external_freqs) |
| void | save_topology_space_to_trprobs_file (topology_space tsp, char *filename, double credible) |
| Save topology_space to a file, in format nexus w/ trprobs, up to "credible" cummul frequency. | |
| int | estimate_treesize_from_file (char *seqfilename) |
| Quickly counts the number of leaves in a tree file, without storing any info. Assumes file and trees are well-formed. | |
| topology_space | new_topology_space (void) |
| Allocates memory for topology_space_struct (set of trees present in nexus file). | |
| void | del_topology_space (topology_space tsp) |
| Free memory from topology_space_struct. | |
Reads tree files in nexus format and creates a vector of topologies.
The topology_space_struct is distinct from the newick_space_struct since all trees here must share same char_vector (newick spaces can have general, uncomparable trees), and also since we store the distribution of trees (that is, each tree will have a frequency/representativity associated to it, as typical from Bayesian analyes).
| void add_string_with_size_to_topology_space | ( | topology_space * | tsp, |
| char * | long_string, | ||
| size_t | string_size, | ||
| bool | use_root_location | ||
| ) |
Read tree in newick format until char string_size, returning updated topolgy_space. Auxiliary for python module.
Read tree in newick format until char string_size, returning updated topolgy_space. Auxiliary for python module.
 1.8.13
1.8.13