 |
biomcmc-lib
0.1
low level library for phylogenetic analysis
|
 |
biomcmc-lib
0.1
low level library for phylogenetic analysis
|
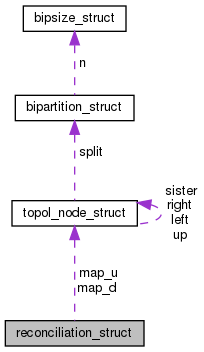
mapping between gene tree nodes (this) and (external) species tree nodes More...
#include <genetree.h>
Data Fields | |
| topol_node * | map_d |
| topol_node * | map_u |
| Mapping of all nodes from gene to species (the first gene::nnodes are fixed) | |
| int * | sp_id |
| Mapping of all nodes from gene to species, assuming gene tree is upside down (unrooted TESTING version) | |
| int * | sp_count |
| mapping of gene (this tolopogy) leaf to ID of taxon in species tree | |
| int | sp_size |
| how many copies of each species are present in this gene (used by deepcoal) | |
| int | size_diff |
| effective number of species present in gene family | |
| int * | dup |
| twice the difference in number of leaves between gene tree and (reduced/effective) species tree | |
| int * | ndup_d |
| indexes of duplication nodes on gene tree, and number of such nodes (unused for now) | |
| int * | ndup_u |
| number of duplications below node (edge above node, since struct assume node == edge above it) | |
| int * | nlos_d |
| number of duplications above node (edge upside down, thus "children" are 'up' and 'sister') | |
| int * | nlos_u |
| number of losses below node and edge above node | |
| int | ndups |
| number of losses above node, including edge above it | |
| int | nloss |
| minimum number of duplications over all possible rootings, acc. to reconciliation_struct::dup | |
| int | ndcos |
| number of losses corresponding to rooting (edge) that minimizes duplications | |
mapping between gene tree nodes (this) and (external) species tree nodes
 1.8.13
1.8.13