 |
biomcmc-lib
0.1
low level library for phylogenetic analysis
|
 |
biomcmc-lib
0.1
low level library for phylogenetic analysis
|
Probability distribution functions and auxiliary mathematical functions from statistical package R. More...
Go to the source code of this file.
Data Structures | |
| struct | discrete_sample_struct |
Typedefs | |
| typedef struct discrete_sample_struct * | discrete_sample |
Functions | |
| void | biomcmc_discrete_gamma (double alpha, double beta, double *rate, int nrates) |
| Ziheng Yang's gamma discretization of rates. | |
| double | biomcmc_dexp_dt (double d, double lambda, double m, bool log_p) |
| pdf of discrete truncated exponential (d is discrete, m is maximum value) More... | |
| double | biomcmc_pexp_dt (double d, double lambda, double m, bool log_p) |
| cdf of discrete truncated exponential (d is discrete, m is maximum value): calculates P(D <= d) | |
| double | biomcmc_qexp_dt (double p, double lambda, double m, bool log_p) |
| quantile of discrete truncated exponential, that is, finds d s.t. P(D <= d) >= p | |
| double | biomcmc_dgamma (double x, double alpha, double beta, bool log_p) |
| gamma density | |
| double | biomcmc_qgamma (double p, double alpha, double beta, bool log_p) |
| gamma quantile (inverse CDF) | |
| double | biomcmc_pgamma (double x, double alpha, double beta, bool log_p) |
computes the cummulative distribution function for the gamma distribution with shape parameter alpha and rate parameter beta, s.t. ![$ E[X] = \alpha/\beta $](form_0.png) . The same as the (lower) incomplete gamma function. . The same as the (lower) incomplete gamma function. | |
| double | biomcmc_dnorm (double x, double mu, double sigma, bool log_p) |
| double | biomcmc_qnorm (double p, double mu, double sigma, bool log_p) |
| double | biomcmc_pnorm (double x, double mu, double sigma, bool log_p) |
| double | biomcmc_dlnorm (double x, double meanlog, double sdlog, bool log_p) |
| double | biomcmc_qlnorm (double p, double meanlog, double sdlog, bool log_p) |
| double | biomcmc_plnorm (double x, double meanlog, double sdlog, bool log_p) |
| double | biomcmc_dpois (double x, double lambda, bool log_p) |
| double | biomcmc_qpois (double p, double lambda, bool log_p) |
| double | biomcmc_ppois (double x, double lambda, bool log_p) |
| double | biomcmc_rng_gamma (double alpha, double beta) |
| double | biomcmc_rng_norm (double mu, double sigma) |
| Returns a random number from a Normal distribution N(mu, sigma^2) using 52 bits of precision. | |
| double | biomcmc_rng_lnorm (double meanlog, double sdlog) |
| double | biomcmc_rng_pois (double mu) |
| double | biomcmc_lgammafn (double x, int *sgn) |
| double | biomcmc_gammafn (double x) |
| double | biomcmc_log1p (double x) |
compute the relative error logarithm 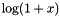 (C99 standard) (C99 standard) | |
| double | biomcmc_log1pmx (double x) |
accurate calculation of 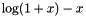 , particularly for small x , particularly for small x | |
| double | biomcmc_expm1 (double x) |
compute 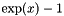 accurately also when x is close to zero, i.e. accurately also when x is close to zero, i.e. 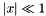 | |
| discrete_sample | new_discrete_sample_from_frequencies (double *prob, size_t size) |
| void | del_discrete_sample (discrete_sample g) |
| size_t | biomcmc_rng_discrete (discrete_sample g) |
| double | biomcmc_discrete_sample_pdf (discrete_sample g, size_t k) |
| double | biomcmc_logspace_add (double logx, double logy) |
| double | biomcmc_logspace_sub (double logx, double logy) |
| bool | biomcmc_isfinite (double x) |
| check if number is between minus infinity and plus infinity, or NaN | |
Probability distribution functions and auxiliary mathematical functions from statistical package R.
Code derived from the R project for Statistical Computing version 2.9.1, available under the GPL license. It might be possible to use directly the standalone mathematical library "Rmath.h" from the R project instead of our implementation. The advantage would be a library updated more often than mine, at the cost of delegating to the guenomu user the installation and maintenance of the extra libraries (like GSL, for instance). In Debian this library can be installed through the package "r-mathlib". The original R library checks for several built-in compiler functions (like log1p(e) for calculating log(e+1) ) but I simply assume the compiler has none and reimplement them. The CDFs always assume the lower tail (upper tails must use 1. - lower tail) or equivalent.
The code for the discrete sampling comes from the GNU Scientific Library version 1.14
| double biomcmc_dexp_dt | ( | double | d, |
| double | lambda, | ||
| double | m, | ||
| bool | log_p | ||
| ) |
pdf of discrete truncated exponential (d is discrete, m is maximum value)
pdf of discrete truncated exponential (d is discrete, m is maximum value)
 1.8.13
1.8.13