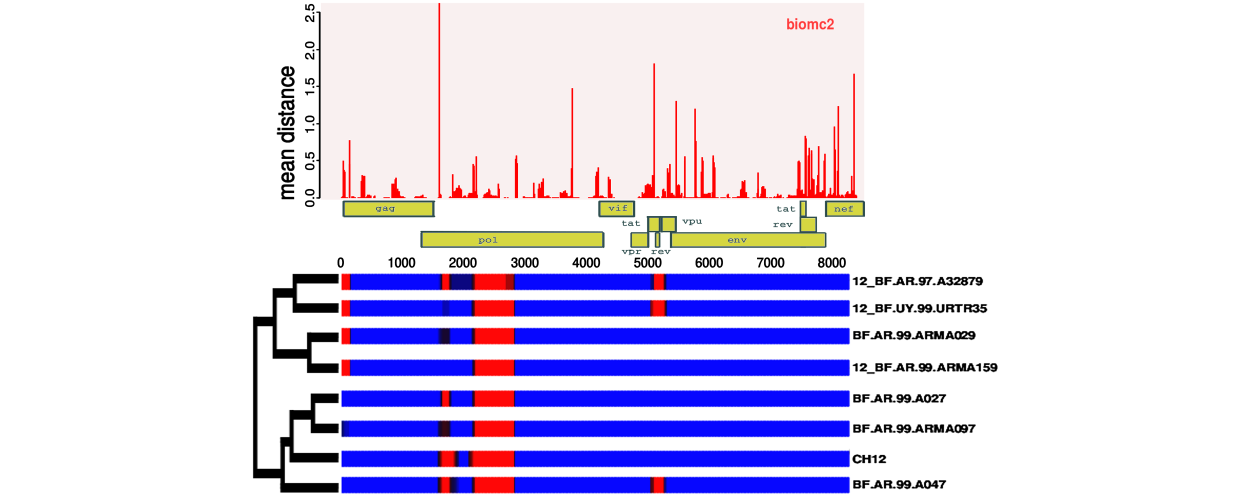
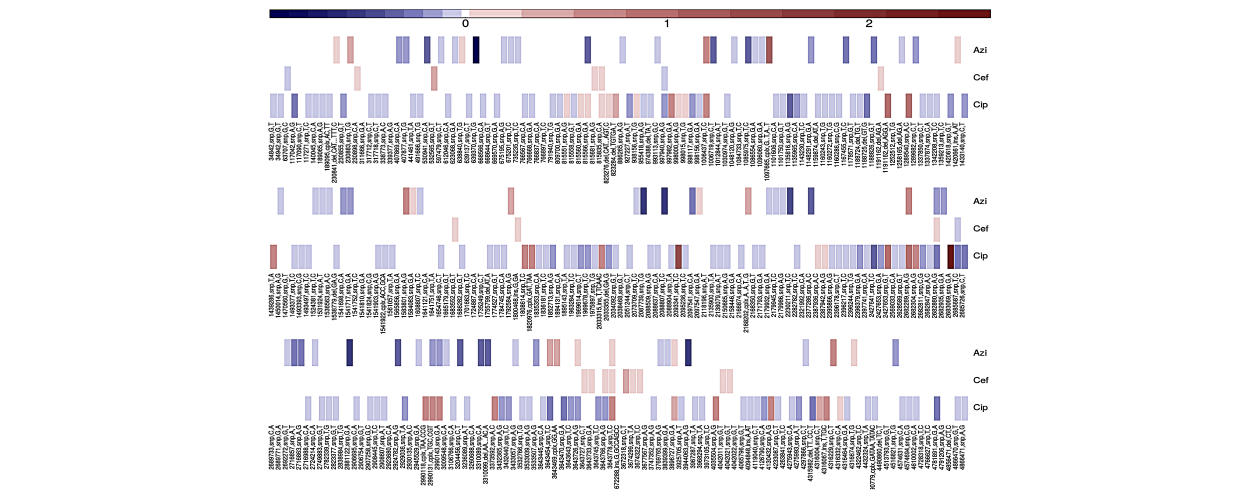
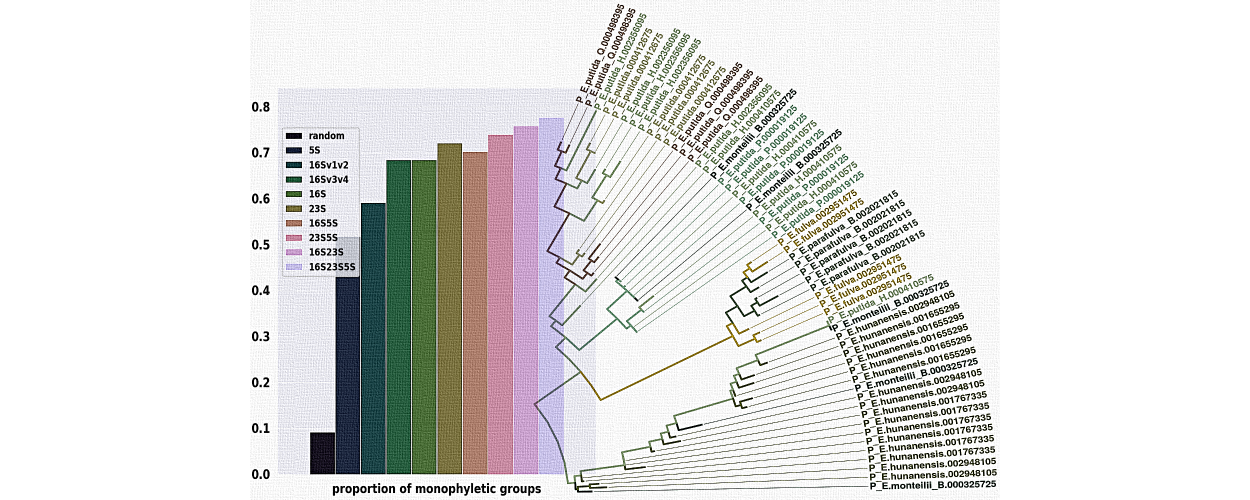
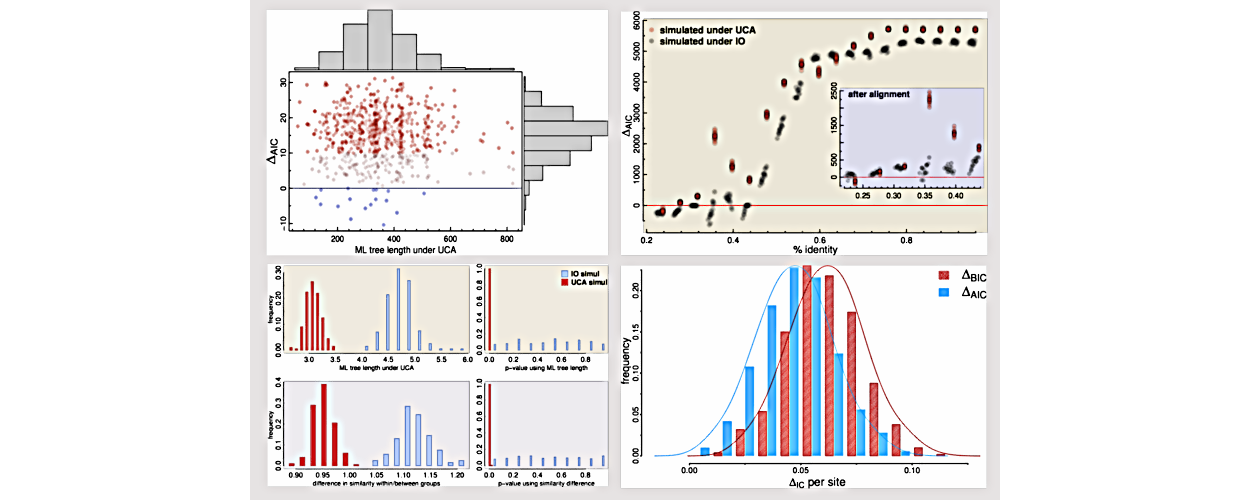
Below you will find links to other pages with code, documentation, and technical notes for bioinformatics and computational evolutionary biology software.
Software and code
Super Distance
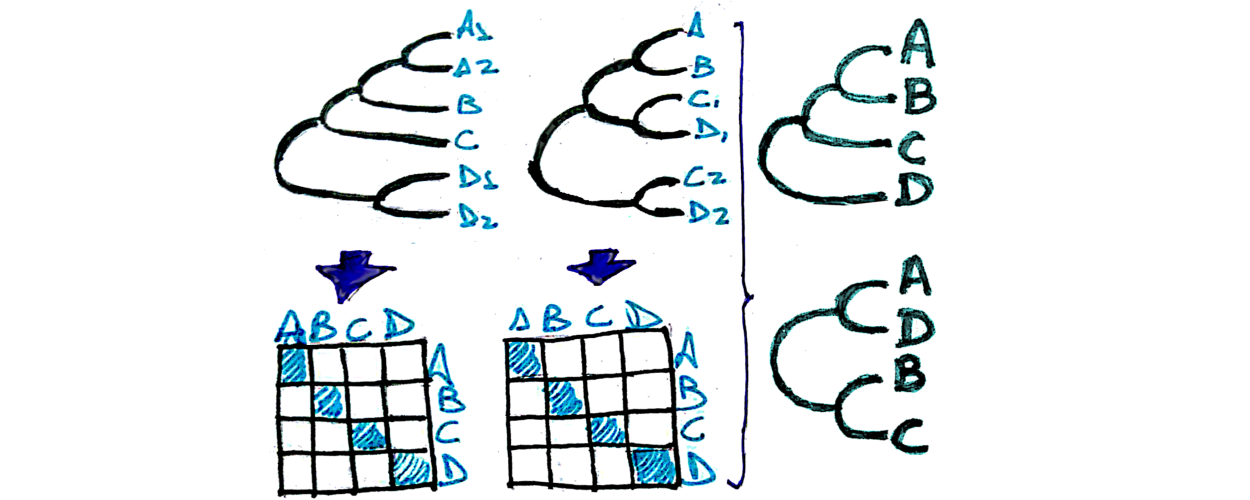
Supertree estimation from gene family trees, using matrix representation with distances
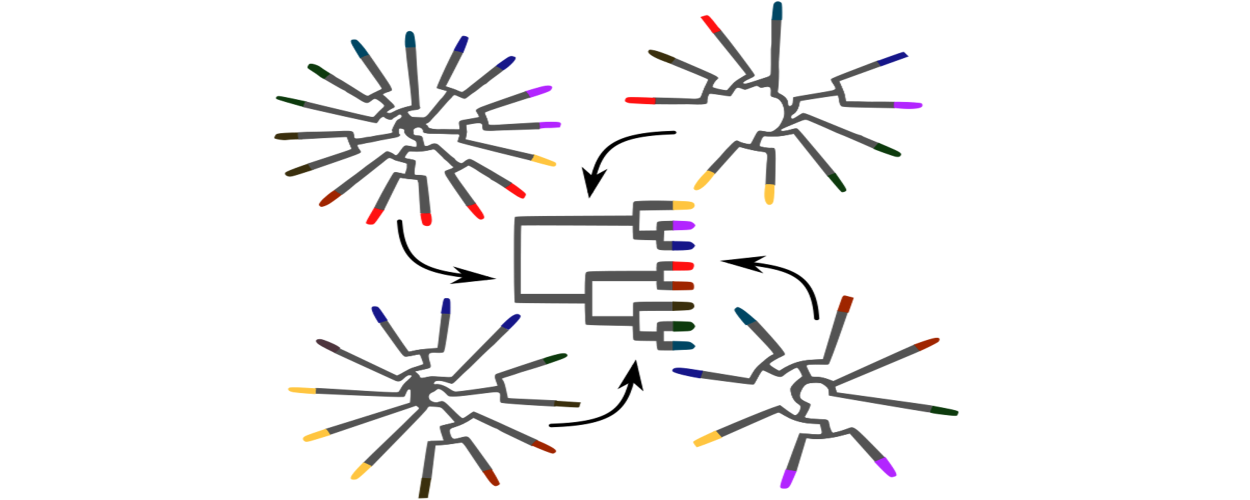
Guenomu
Hierarchical Bayesian species tree estimation from multi-gene family data
Genefam-dist
Signature inference of gene family trees from anchoring species trees.
biomcmc-lib library
C library with low level functions for phylogenetic analyses
Curupixa
(Experimental) thread-safe API-aware C library to supercede biomcmc-lib someday
Supplementary analyses for manuscripts
Salmonella Biofilm
Jupyter notebooks with analyses of experimental evolution of Salmonella biofilms
Documentation and Texts
Binfie notes
Blog with technical notes on bioinformatics algorithms and implementations